Predict
Predict
# Enter the website
Visit Predict (opens new window) to enter our platform page.
# Input your data
# 1 Input Peptide
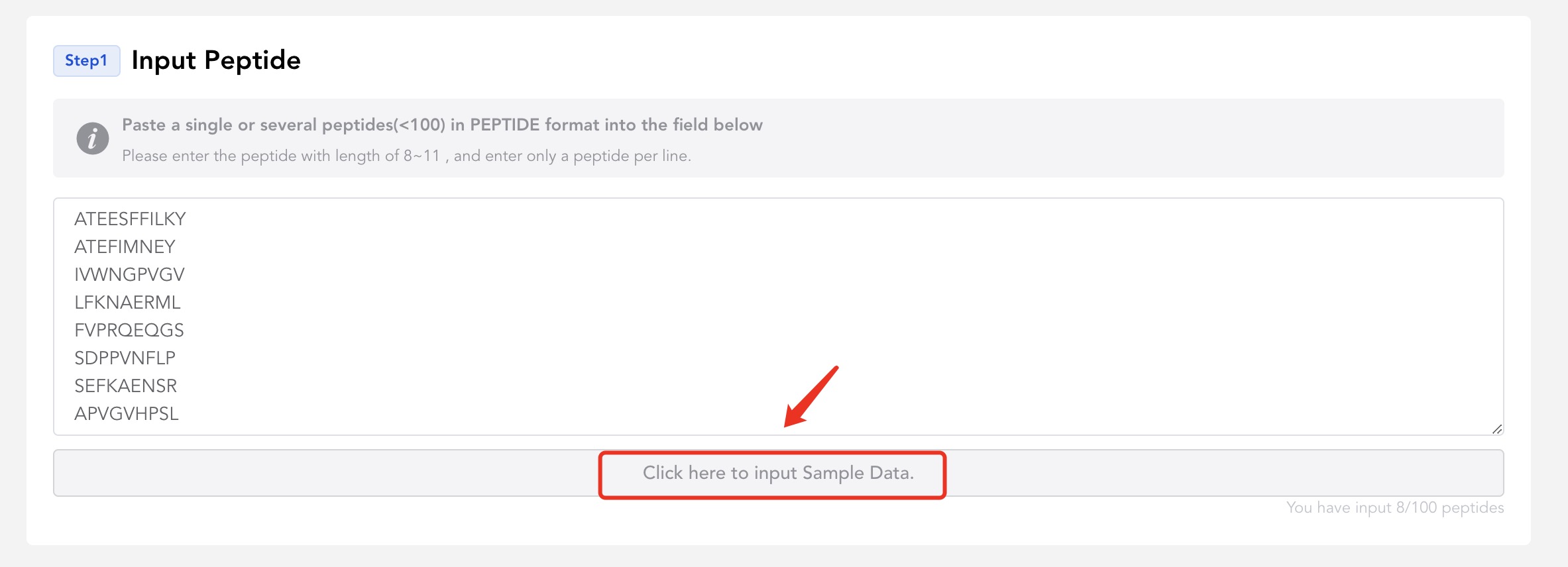
You should enter the peptide with length of 8~11 , and enter only a peptide per line.But you can enter up to 100 sequences.
- Of course, you can also click this button to enter our default sample for testing.
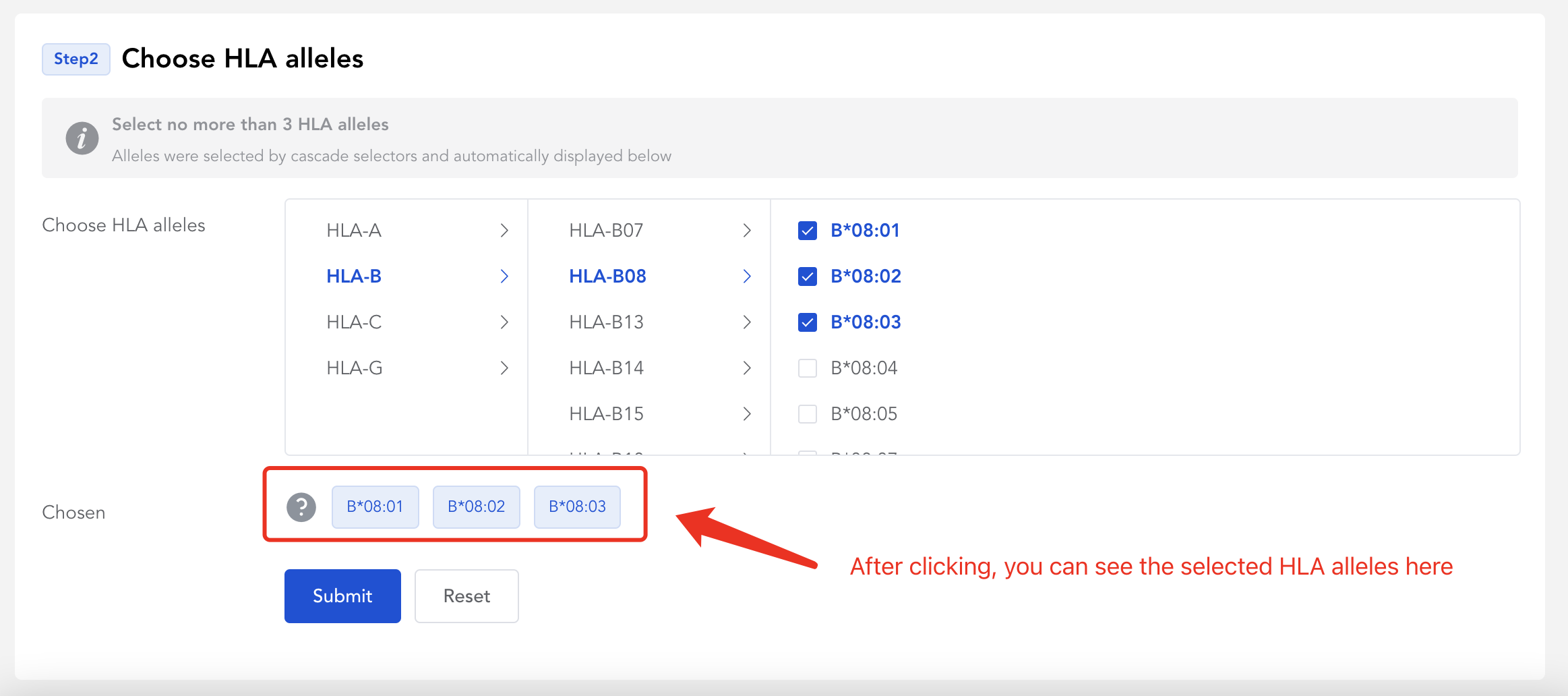
# 2 Choose HLA alleles
You should select no more than 3 HLA alleles.
# Calculate Result
# 1 Result Table
After clicking "submit", you can see such a table.
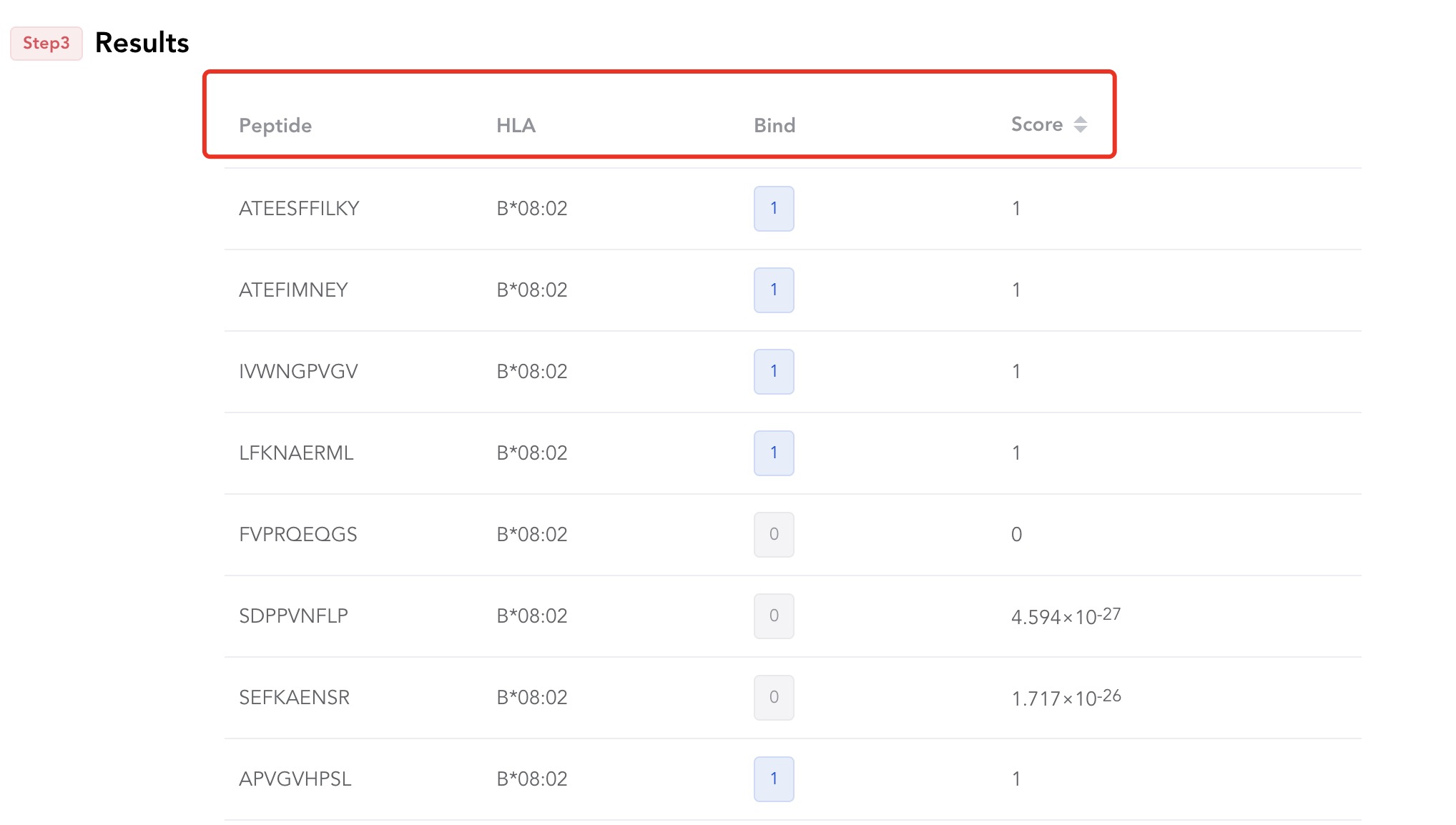
| Columns | Description |
|---|---|
| Peptide | inputted |
| MHC | inputted |
| Bind | Whether the peptide binds to MHC (1 indicates binding, 0 indicates non binding) |
| Score | Binding fraction of peptide to MHC (after softmax) |
# 2 Download
There is a "download" button at the bottom of the table. You can click this button and the download page will pop up.
click button
download